Coordinates on the sky and the chip¶
In every marx simulation, one or more sources are placed at some sky position. marx simulates photons coming from that position, traces them through the mirror and gratings and finally places them on the chip. With a known aspect solution, chip coordinates can then be transformed back to sky coordinates. In general, this will not recover the exact sky position where a photon started out. A big part of that is scatter in the mirrors, which blurs the image. However, with a large number of photons, we can fit the average position which should be close to the real sky position.
In real observations, other factors contribute, such as the finite
resolution of the detectors (marx usually takes that into account, but it can
be switched off through the --pixadj="EXACT" switch in marx2fits)
and the uncertainty of the aspect solution.
Within a single observation, positions will be less certain for fainter sources (due to Poisson statistics) and for sources at a larger off-axis angles (due to the larger PSF).
Chandra Orion Ultradeep project¶
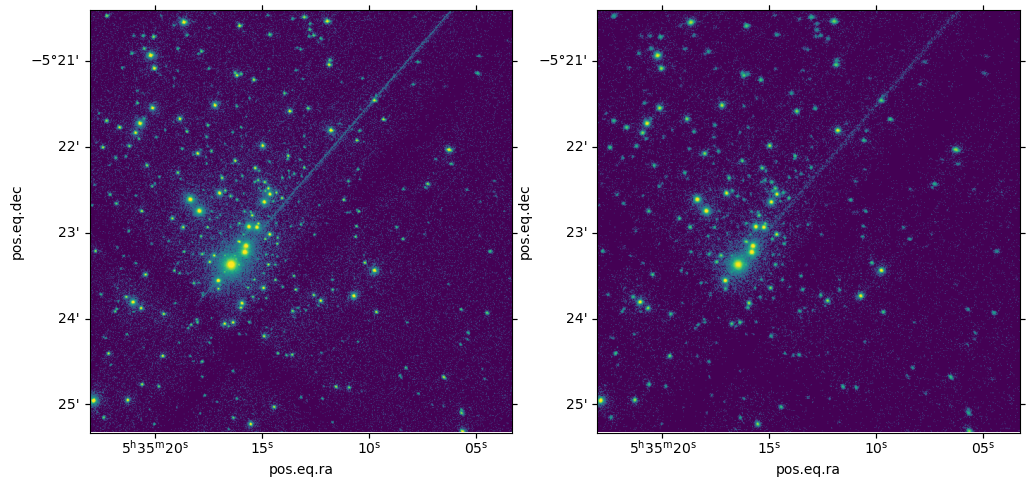
The Orion Nebula Cluster (ONC) is a dense star forming region with about 1600 X-ray sources observed in the COUP survey by Getman et al (2005). We simulate this field with marx and then run a source detection to check how well we recover the input coordinates. This will depend on the number of counts detected and the position in the field. To simplify the simulation input, we assume that all sources have flat lightcurves and are monoenergetic at the observed mean energy (the energy matters because the effective area is energy dependent and so is the PSF). We write a short C code that reads an input coordinate list and generates the photons in this manner.
In the observation, the brightest sources are piled-up. We don’t bother simulating this here, so for the visualization we just set the scaling limits to bring out the fainter details and ignore the bright peaks.
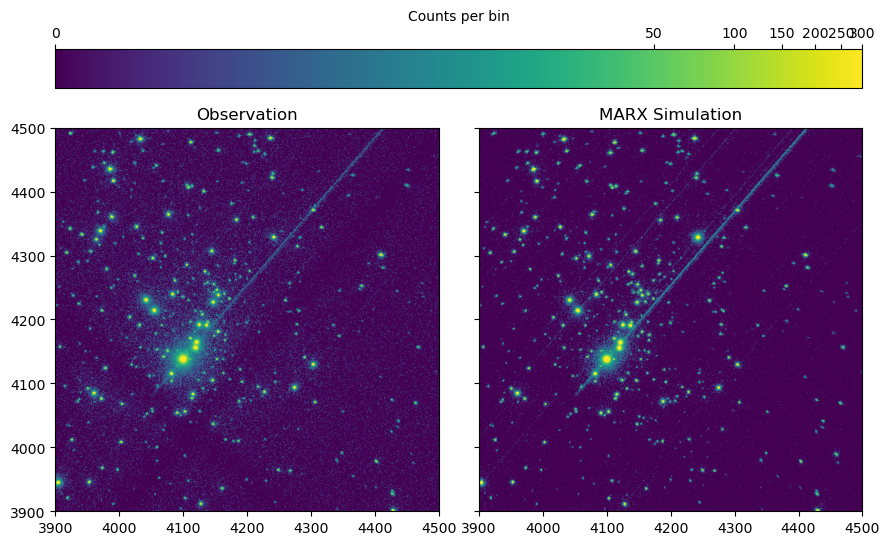
Image of the observed data (left) and simulation (right). The diagonal line from the center to the top right is the read-out streak from :math:\theta^1 Ori C.
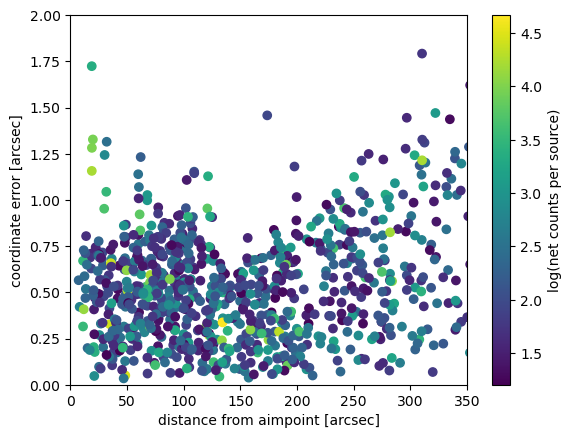
Apart from a clump close to the aimpoint (where sources in the ONC are so close to each other that they can easily be confused), the distribution of coordinate errors spreads out with increasing distance, i.e. size of the PSF.
Simulations with an IMAGE source¶
The SourceType=IMAGE in marx is a strange beast that behaves in ways that users might find surprising. When using this source type, marx will read in an image file and use the pixel values as relative probabilities for generating photons across the image.
Marx will read the WCS in the image header, but only use the absolute value for the pixel size, not the orientation or position. The center of the image is always supposed to be at the nominal pointing position and the image x/y axes are always aligned with the sky coordinate system in marx.
Regular Grid (ACIS)¶
In this example we place a radial grid of sources on the sky. Each source emits an equal number of photons (exactly, no Poisson statistics) so that we can compare the accuracy of the position we recover. Note that the detected number of photons will be smaller for off-axis photons because of vignetting!
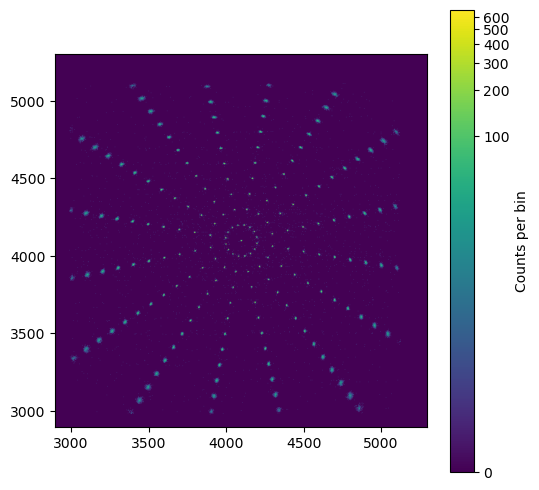
Image of the simulation. The size of the PSF increases further away from the aimpoint.
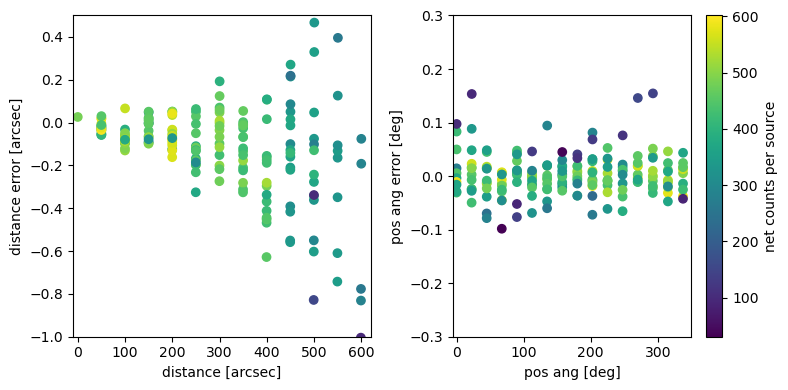
left: The error in the position (measured radially to the optical axis) increases with the distance to the optical axis. One part of this is just that the effective area and thus the number of counts decreases. There is also a systematic trend where sources at larger off-axis angle are systematically fitted too close to the center. Further investigation is necessary to check if this is a problem of marx or the CIAO tool celldetect. In any case, the typical offset is below 0.2 arcsec, which is less than half a pixel in ACIS. right: Difference in position angle between input and fit.
The input position is typically recovered to much better than one pixel for sources with a few hundred counts. There is a small systematic trend that needs to be studied further.
Regular grid (HRC)¶
Same as above, but with HRC-I as a detector.
The field-of-view for the HRC-I is larger for than for ACIS-I, but the PSF becomes very large at large off-axis angles and thus the positional uncertainty will be so large that a comparison to marx is no longer helpful to test the accuracy of the marx simulations.
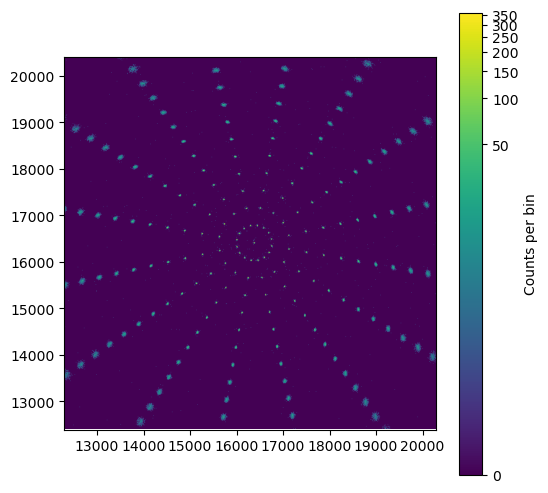
Image of the simulation. The size of the PSF increases further away from the aimpoint.
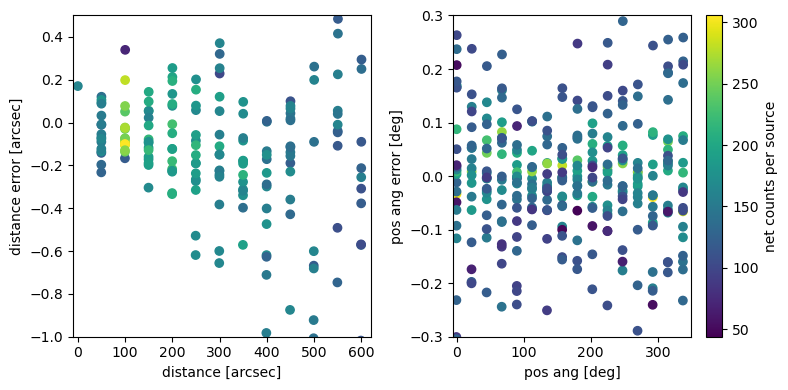
See previous example. The same trends are visible with a slightly larger scatter.
In the central few arcmin the input position is typically recovered to better than 0.2 pixels for sources with a few hundred counts.
